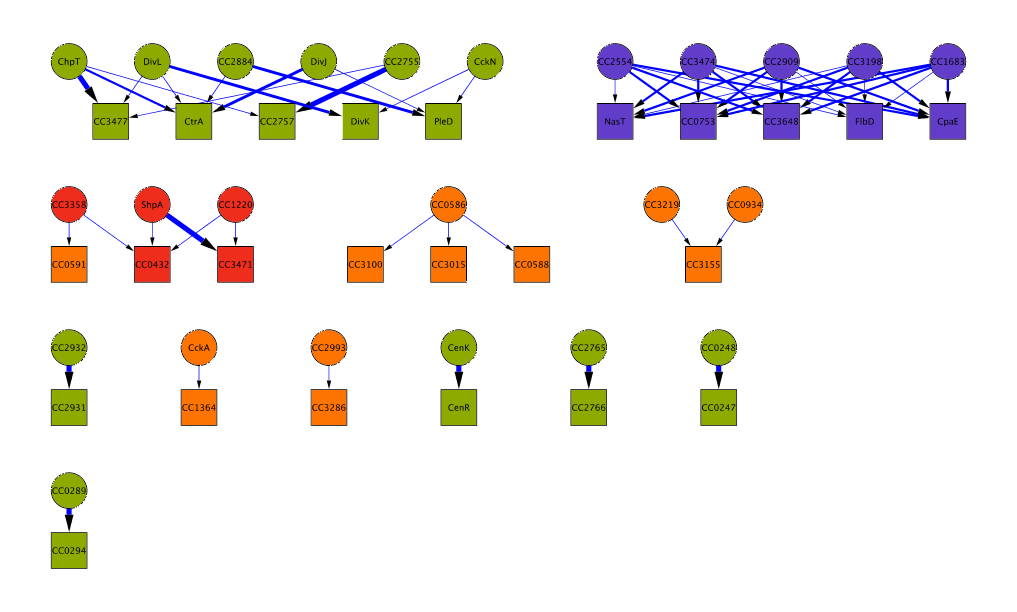
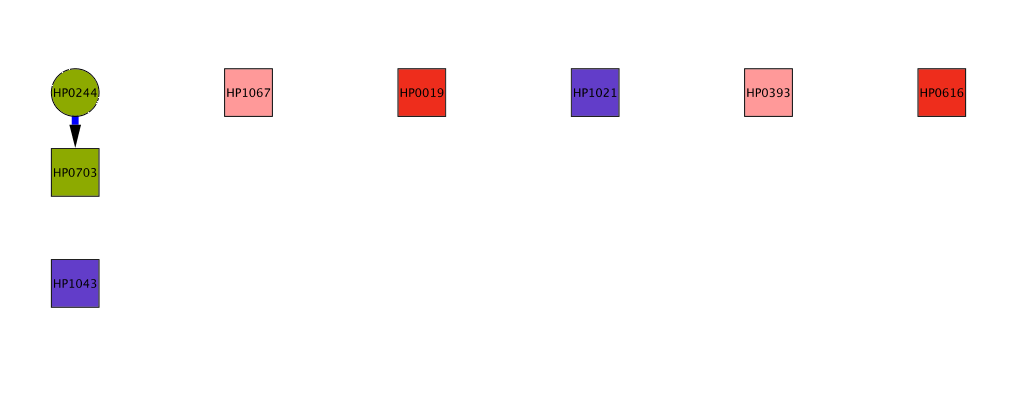
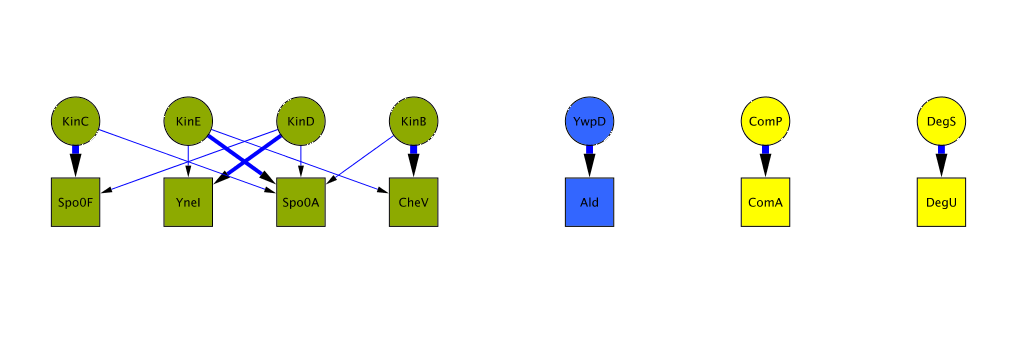
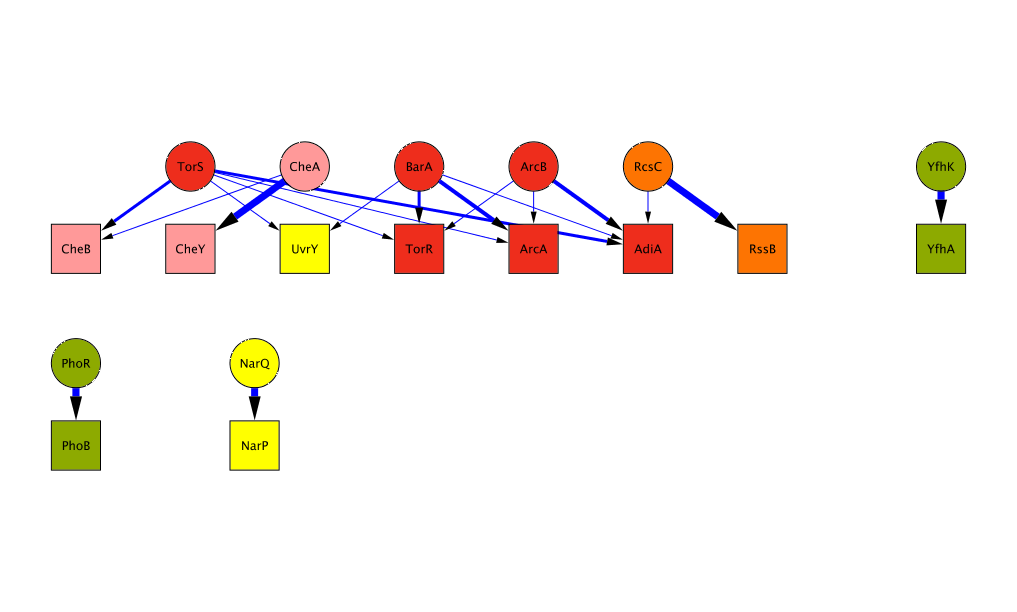
The Bayesian algorithm we developed expicitly models the amino acid composition of interacting kinase/regulator pairs to predict interaction specificity of orphan kinases and regulators across all sequenced bacterial genomes. Kinases and regulators come in different families of varying size. We are most confident about predictions involving kinases of the largest subfamilies, such as HisKA or H3. On this website, you can either look at the predictions for your bacterium of choice or look at a few examples of predicted interaction networks. The complete list of predictions can be downloaded from
the publisher's website.
Accurate prediction of protein-protein interactions from sequence alignments using a Bayesian method
Lukas Burger and Erik van Nimwegen
Molecular Systems Biology 4:165